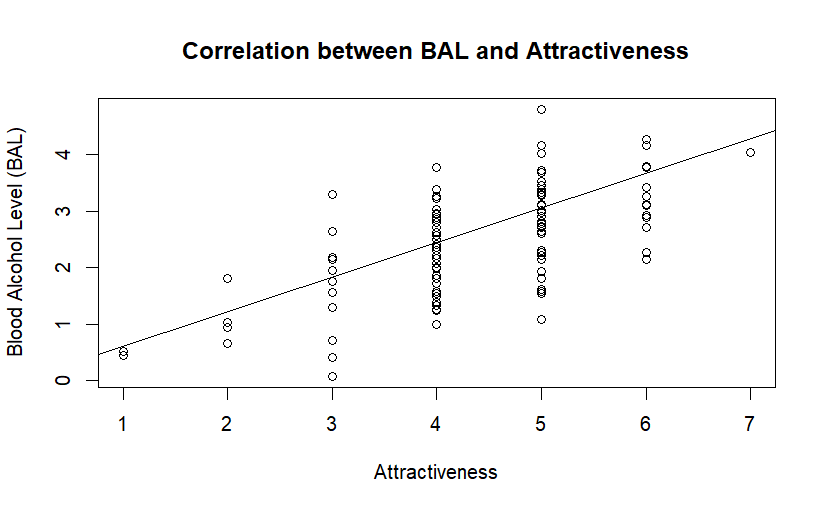
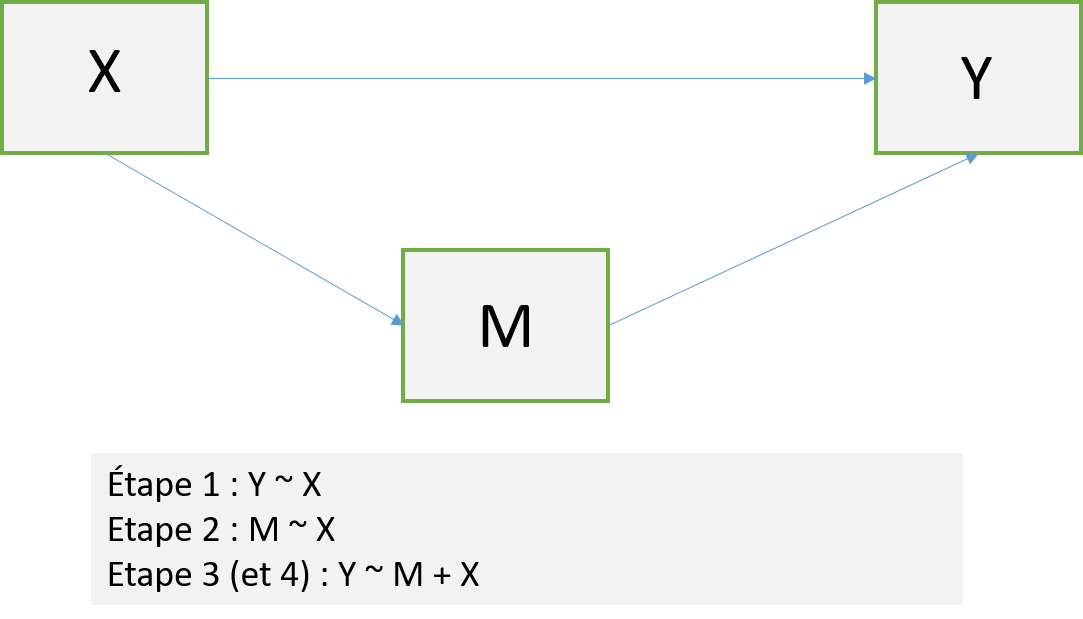
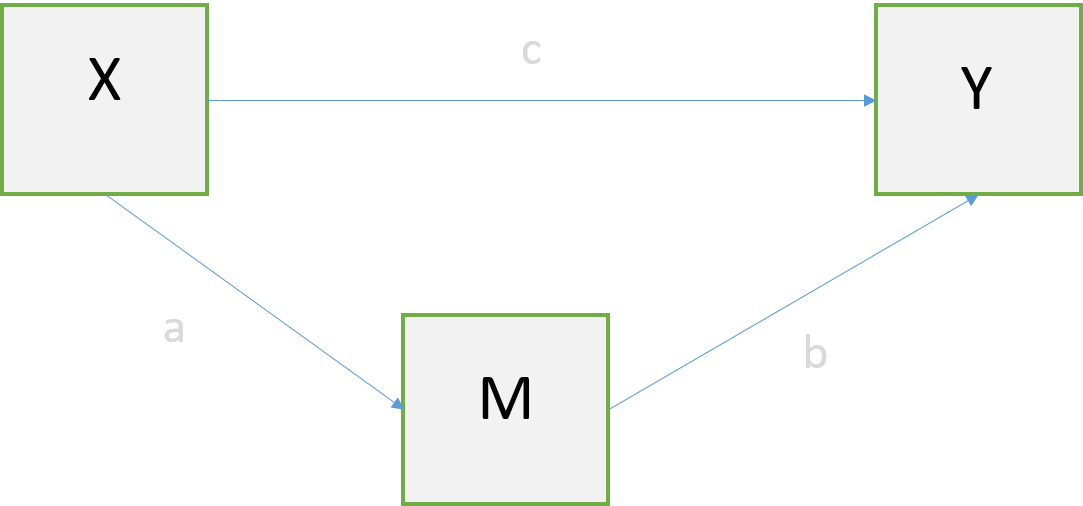
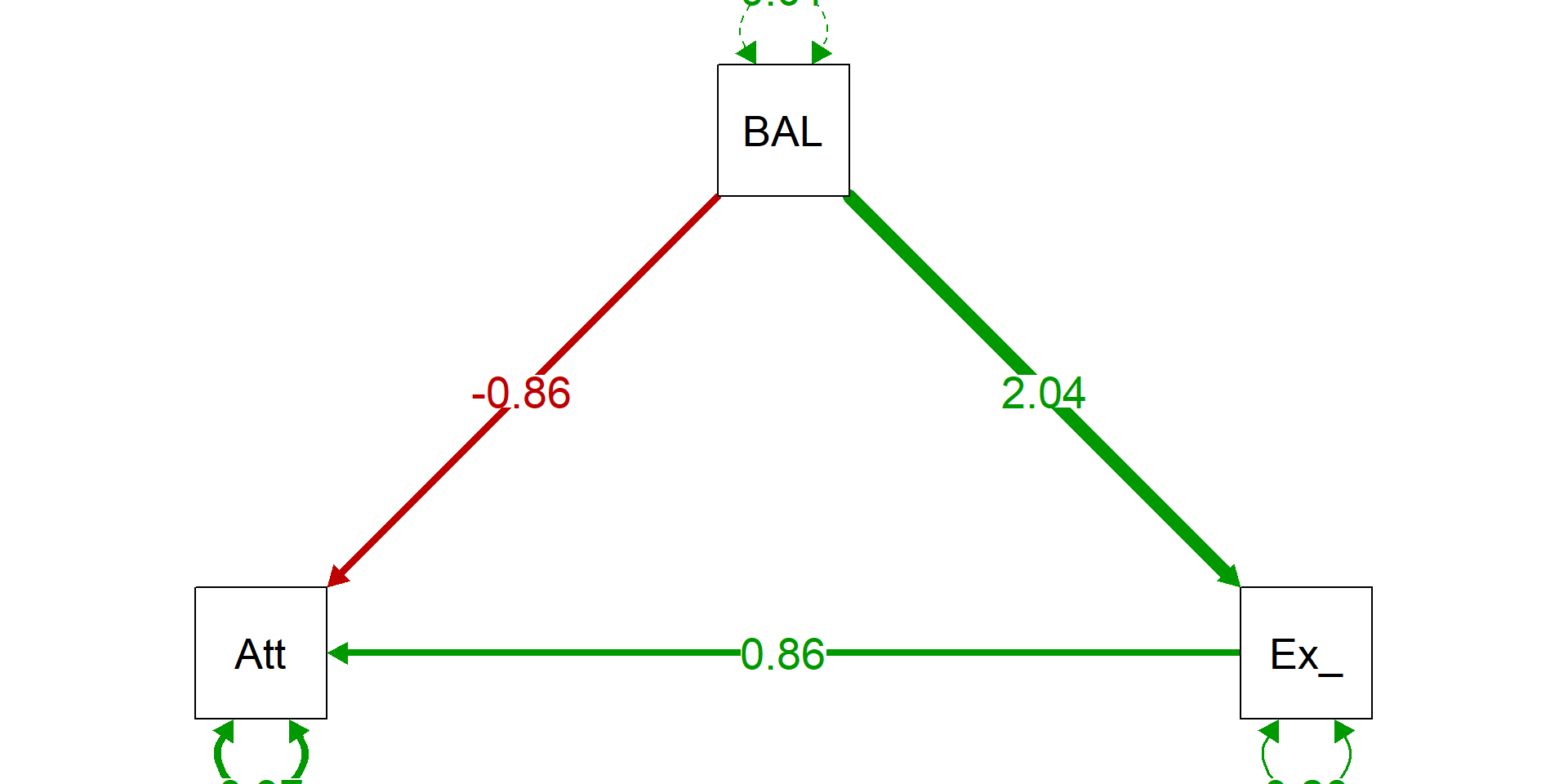
attach(datRPSBSp) # Attacher car R refuse les $
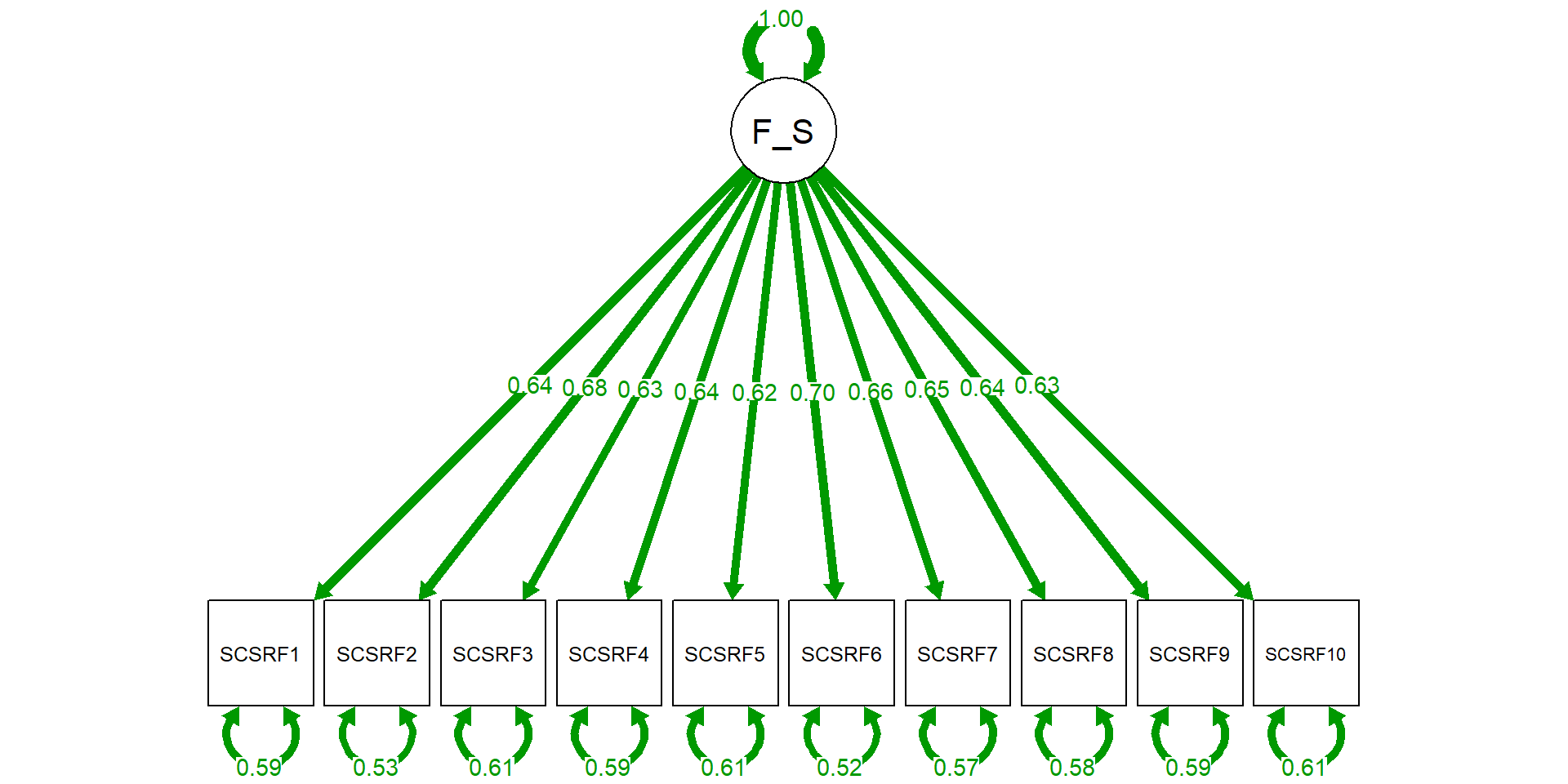
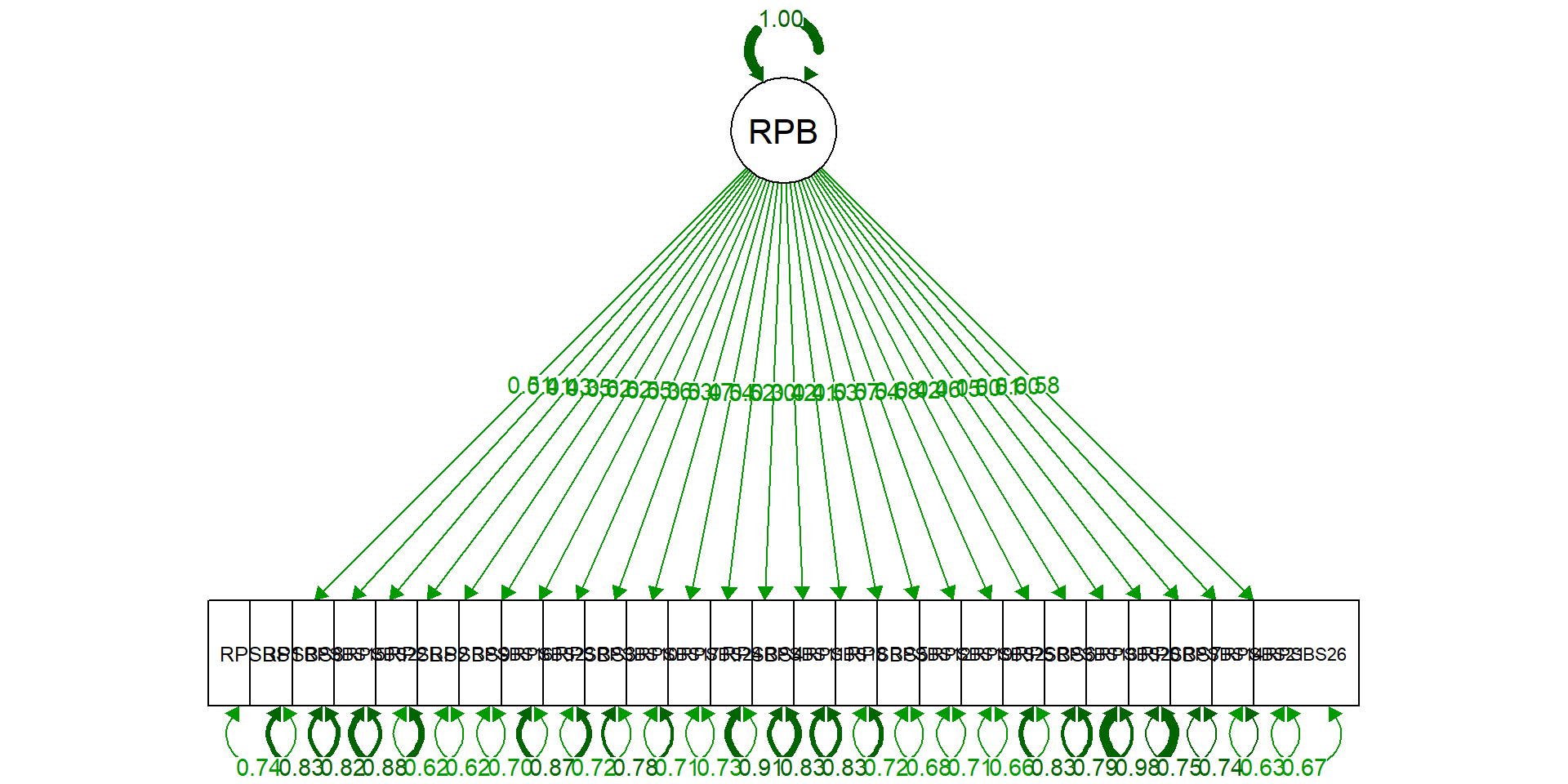
modelR <- 'RPBS =~ RPSBS1 + RPSBS8 + RPSBS15 + RPSBS22
+ RPSBS2 + RPSBS9 + RPSBS16 + RPSBS23
+ RPSBS3 + RPSBS10 + RPSBS17 + RPSBS24
+ RPSBS4 + RPSBS11 + RPSBS18
+ RPSBS5 + RPSBS12 + RPSBS19 + RPSBS25
+ RPSBS6 + RPSBS13 + RPSBS20
+ RPSBS7 + RPSBS14 + RPSBS21 + RPSBS26'
fitMLRR <- cfa (modelR, std.lv=T, estimator="MLR", data=datRPSBSp)
summary(fitMLRR, standardized=T, modindices = F, fit.measures=T)
lavaan 0.6.16 ended normally after 13 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 52
Number of observations 744
Model Test User Model:
Standard Scaled
Test Statistic 287.168 289.799
Degrees of freedom 299 299
P-value (Chi-square) 0.678 0.638
Scaling correction factor 0.991
Yuan-Bentler correction (Mplus variant)
Model Test Baseline Model:
Test statistic 4080.816 4114.998
Degrees of freedom 325 325
P-value 0.000 0.000
Scaling correction factor 0.992
User Model versus Baseline Model:
Comparative Fit Index (CFI) 1.000 1.000
Tucker-Lewis Index (TLI) 1.003 1.003
Robust Comparative Fit Index (CFI) 1.000
Robust Tucker-Lewis Index (TLI) 1.003
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -25420.834 -25420.834
Scaling correction factor 1.002
for the MLR correction
Loglikelihood unrestricted model (H1) -25277.250 -25277.250
Scaling correction factor 0.993
for the MLR correction
Akaike (AIC) 50945.669 50945.669
Bayesian (BIC) 51185.495 51185.495
Sample-size adjusted Bayesian (SABIC) 51020.375 51020.375
Root Mean Square Error of Approximation:
RMSEA 0.000 0.000
90 Percent confidence interval - lower 0.000 0.000
90 Percent confidence interval - upper 0.012 0.012
P-value H_0: RMSEA <= 0.050 1.000 1.000
P-value H_0: RMSEA >= 0.080 0.000 0.000
Robust RMSEA 0.000
90 Percent confidence interval - lower 0.000
90 Percent confidence interval - upper 0.012
P-value H_0: Robust RMSEA <= 0.050 1.000
P-value H_0: Robust RMSEA >= 0.080 0.000
Standardized Root Mean Square Residual:
SRMR 0.025 0.025
Parameter Estimates:
Standard errors Sandwich
Information bread Observed
Observed information based on Hessian
Latent Variables:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
RPBS =~
RPSBS1 0.514 0.037 13.815 0.000 0.514 0.514
RPSBS8 0.405 0.035 11.703 0.000 0.405 0.408
RPSBS15 0.426 0.037 11.624 0.000 0.426 0.429
RPSBS22 0.349 0.036 9.781 0.000 0.349 0.351
RPSBS2 0.607 0.033 18.338 0.000 0.607 0.616
RPSBS9 0.617 0.036 17.266 0.000 0.617 0.619
RPSBS16 0.545 0.035 15.387 0.000 0.545 0.548
RPSBS23 0.364 0.036 10.121 0.000 0.364 0.364
RPSBS3 0.530 0.036 14.871 0.000 0.530 0.533
RPSBS10 0.466 0.037 12.562 0.000 0.466 0.469
RPSBS17 0.527 0.035 15.207 0.000 0.527 0.537
RPSBS24 0.518 0.035 14.831 0.000 0.518 0.523
RPSBS4 0.298 0.035 8.412 0.000 0.298 0.298
RPSBS11 0.407 0.035 11.562 0.000 0.407 0.418
RPSBS18 0.408 0.036 11.429 0.000 0.408 0.414
RPSBS5 0.533 0.035 15.069 0.000 0.533 0.533
RPSBS12 0.566 0.038 14.753 0.000 0.566 0.567
RPSBS19 0.535 0.034 15.870 0.000 0.535 0.539
RPSBS25 0.578 0.037 15.540 0.000 0.578 0.582
RPSBS6 0.414 0.039 10.593 0.000 0.414 0.416
RPSBS13 0.454 0.034 13.199 0.000 0.454 0.457
RPSBS20 0.154 0.039 3.978 0.000 0.154 0.154
RPSBS7 0.498 0.036 13.804 0.000 0.498 0.501
RPSBS14 0.511 0.036 14.071 0.000 0.511 0.512
RPSBS21 0.597 0.035 17.294 0.000 0.597 0.604
RPSBS26 0.576 0.034 16.901 0.000 0.576 0.577
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.RPSBS1 0.733 0.039 18.581 0.000 0.733 0.735
.RPSBS8 0.824 0.049 16.898 0.000 0.824 0.834
.RPSBS15 0.804 0.040 19.970 0.000 0.804 0.816
.RPSBS22 0.868 0.041 20.912 0.000 0.868 0.877
.RPSBS2 0.604 0.034 18.013 0.000 0.604 0.621
.RPSBS9 0.612 0.035 17.491 0.000 0.612 0.617
.RPSBS16 0.693 0.038 18.176 0.000 0.693 0.700
.RPSBS23 0.866 0.044 19.842 0.000 0.866 0.867
.RPSBS3 0.706 0.038 18.720 0.000 0.706 0.716
.RPSBS10 0.770 0.040 19.428 0.000 0.770 0.780
.RPSBS17 0.686 0.034 20.436 0.000 0.686 0.712
.RPSBS24 0.713 0.038 18.982 0.000 0.713 0.727
.RPSBS4 0.914 0.044 20.690 0.000 0.914 0.911
.RPSBS11 0.783 0.043 18.285 0.000 0.783 0.825
.RPSBS18 0.807 0.045 17.954 0.000 0.807 0.829
.RPSBS5 0.714 0.038 18.604 0.000 0.714 0.715
.RPSBS12 0.677 0.039 17.348 0.000 0.677 0.679
.RPSBS19 0.698 0.039 17.878 0.000 0.698 0.709
.RPSBS25 0.652 0.037 17.751 0.000 0.652 0.661
.RPSBS6 0.817 0.045 18.332 0.000 0.817 0.827
.RPSBS13 0.784 0.039 20.177 0.000 0.784 0.792
.RPSBS20 0.972 0.051 18.894 0.000 0.972 0.976
.RPSBS7 0.741 0.042 17.556 0.000 0.741 0.749
.RPSBS14 0.735 0.040 18.235 0.000 0.735 0.737
.RPSBS21 0.619 0.035 17.519 0.000 0.619 0.635
.RPSBS26 0.664 0.037 17.774 0.000 0.664 0.667
RPBS 1.000 1.000 1.000
resid(fitMLRR,type="standardized") # Saturations standardisées
$type
[1] "standardized"
$cov
RPSBS1 RPSBS8 RPSBS15 RPSBS22 RPSBS2 RPSBS9 RPSBS16 RPSBS23 RPSBS3
RPSBS1 0.000
RPSBS8 -0.247 0.000
RPSBS15 -1.224 -2.044 0.000
RPSBS22 -0.049 -0.314 0.995 0.000
RPSBS2 -0.070 0.650 0.840 0.523 0.000
RPSBS9 0.626 0.023 -1.165 -1.664 -1.414 0.000
RPSBS16 -0.166 0.648 -0.343 2.030 -0.104 0.960 0.000
RPSBS23 1.161 0.086 1.932 -0.719 -0.027 -0.214 1.177 0.000
RPSBS3 0.101 0.885 -0.573 0.127 -0.685 1.675 0.286 -1.786 0.000
RPSBS10 -0.461 -0.148 -0.051 -0.810 -0.272 2.121 -0.060 0.567 -1.079
RPSBS17 0.126 -1.688 -0.070 -0.935 -0.695 -0.477 1.790 1.798 0.679
RPSBS24 -0.107 1.514 1.421 -1.865 -1.541 0.599 -1.172 0.876 -0.508
RPSBS4 0.127 -0.431 -1.236 1.547 -0.413 -0.529 -1.127 -0.141 1.251
RPSBS11 2.180 0.661 -0.935 -0.159 0.705 0.256 -1.121 0.212 1.283
RPSBS18 -0.451 -0.214 0.108 -0.539 0.035 0.767 -0.235 -2.493 0.185
RPSBS5 1.322 -1.049 -0.284 -0.206 1.835 -0.119 0.568 -0.647 -0.443
RPSBS12 -0.438 0.797 1.992 0.386 -0.168 -0.090 0.723 -0.365 -0.159
RPSBS19 -0.881 1.835 -1.085 1.249 -1.834 0.420 1.790 0.751 0.883
RPSBS25 0.081 0.621 -0.297 -0.331 0.425 0.190 0.038 -1.210 -1.237
RPSBS6 1.073 0.559 1.286 0.287 1.331 -1.366 -0.042 -0.798 -0.911
RPSBS13 -0.823 -1.081 -1.488 -0.239 0.274 -1.310 0.079 0.458 -1.250
RPSBS20 0.179 -0.115 -1.004 0.291 -0.869 0.757 0.017 -0.507 1.031
RPSBS7 -1.092 0.063 0.872 -1.484 -0.752 0.108 -0.408 0.746 0.451
RPSBS14 -1.416 -2.129 -0.072 0.748 -0.503 1.736 -0.539 -0.598 0.001
RPSBS21 -1.165 0.894 2.162 0.948 0.752 -1.444 -2.178 -1.102 -1.100
RPSBS26 2.019 -1.307 -1.279 0.234 1.864 -0.711 -1.948 0.518 1.701
RPSBS10 RPSBS17 RPSBS24 RPSBS4 RPSBS11 RPSBS18 RPSBS5 RPSBS12 RPSBS19
RPSBS1
RPSBS8
RPSBS15
RPSBS22
RPSBS2
RPSBS9
RPSBS16
RPSBS23
RPSBS3
RPSBS10 0.000
RPSBS17 -0.031 0.000
RPSBS24 0.450 -0.701 0.000
RPSBS4 0.545 1.396 0.793 0.000
RPSBS11 -1.546 0.417 -1.403 1.240 0.000
RPSBS18 -1.126 -0.299 -0.329 0.455 -1.046 0.000
RPSBS5 -0.867 -0.647 1.226 -3.542 -0.337 -0.779 0.000
RPSBS12 1.268 -0.761 -0.369 0.302 0.941 -0.809 -2.064 0.000
RPSBS19 0.276 -0.366 0.818 1.570 -0.625 0.709 -0.905 0.000 0.000
RPSBS25 0.467 0.768 -1.381 -0.180 -0.209 0.891 0.198 0.944 -0.400
RPSBS6 1.286 -0.262 -0.535 1.489 -0.436 0.401 0.587 -0.698 -0.496
RPSBS13 -2.329 0.816 1.276 -0.242 -1.026 0.241 1.747 -0.553 -0.113
RPSBS20 0.007 -0.170 -0.857 0.383 0.880 -0.704 0.936 0.313 -0.872
RPSBS7 -0.356 0.740 -0.621 -1.316 0.786 0.512 0.534 0.642 0.871
RPSBS14 0.269 -0.062 1.678 1.286 -0.006 0.972 0.420 -0.844 -0.001
RPSBS21 0.092 -0.719 2.329 -0.757 -0.357 1.243 0.004 0.957 -0.587
RPSBS26 0.352 0.138 -1.694 -0.752 -0.144 0.329 0.803 -1.216 -1.373
RPSBS25 RPSBS6 RPSBS13 RPSBS20 RPSBS7 RPSBS14 RPSBS21 RPSBS26
RPSBS1
RPSBS8
RPSBS15
RPSBS22
RPSBS2
RPSBS9
RPSBS16
RPSBS23
RPSBS3
RPSBS10
RPSBS17
RPSBS24
RPSBS4
RPSBS11
RPSBS18
RPSBS5
RPSBS12
RPSBS19
RPSBS25 0.000
RPSBS6 -0.740 0.000
RPSBS13 -1.107 1.229 0.000
RPSBS20 0.320 0.278 0.378 0.000
RPSBS7 -0.186 -1.002 -0.306 0.401 0.000
RPSBS14 -0.410 -1.295 0.857 -0.003 0.285 0.000
RPSBS21 1.076 0.239 1.823 -0.397 -1.363 0.158 0.000
RPSBS26 0.473 0.200 1.719 -0.512 1.394 -0.636 -0.726 0.000